Using DBSCAN to Analyze Animal Behavioral Data: A Case Study with Proximity Logger Data
Receiver ID, Sender ID, Timestamp, RSSI
405, 435, 16/05/2024 12:03:43 GPS, -62
405, 425, 16/05/2024 12:03:43 GPS, -85
...Animal behavior analysis is crucial for understanding various aspects of ecology, wildlife management, and conservation efforts. One effective method to analyze such data is by using clustering algorithms, such as DBSCAN (Density-Based Spatial Clustering of Applications with Noise). In this blog post, we will explore how to use DBSCAN to analyze animal behavioral data using proximity logger data. We will provide both Python and R code examples to illustrate the process.
What is DBSCAN?
DBSCAN is a clustering algorithm that groups together points that are closely packed together, marking points that lie alone in low-density regions as outliers. It is particularly useful for discovering clusters of varying shapes and sizes and is robust to noise and outliers.
Proximity Logger Data
Our example proximity logger data gathered by IoSA’s ProxLogs system contains the following columns:
- Receiver ID: The ID of the receiver logger.
- Sender ID: The ID of the sender logger.
- Timestamp: The date and time when the signal was received.
- RSSI: The Received Signal Strength Indicator, which gives an indication of the distance between the sender and receiver.
Here is a sample of the data:
Receiver ID, Sender ID, Timestamp, RSSI
405, 435, 16/05/2024 12:03:43 GPS, -62
405, 425, 16/05/2024 12:03:43 GPS, -85
...Data Preparation
We will start by loading and preparing the data for analysis.
Python Example
import pandas as pd
import numpy as np
from sklearn.cluster import DBSCAN
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.widgets import Slider
# Load the data
data = pd.read_csv('proximity_logger_data.csv')
data['Timestamp'] = pd.to_datetime(data['Timestamp'].str.replace(' GPS', ''), dayfirst=True)
# Filter for relevant animal loggers
animal_loggers = [347, 350, 367, 370, 371, 383, 405, 411, 412, 416, 420, 422, 425, 2010, 2020, 2030, 2040, 2060, 2070, 2110, 2160]
filtered_data = data[data['Receiver ID'].isin(animal_loggers) & data['Sender ID'].isin(animal_loggers)]
# Group data by 15-minute windows
def group_data_by_time_window(df, window='15min'):
df = df.set_index('Timestamp')
grouped = df.groupby([pd.Grouper(freq=window), 'Receiver ID', 'Sender ID']).agg({'RSSI': 'mean'}).reset_index()
return grouped
grouped_data = group_data_by_time_window(filtered_data)R Example
library(dplyr)
library(lubridate)
library(DBI)
library(ggplot2)
# Load the data
data <- read.csv('proximity_logger_data.csv', stringsAsFactors = FALSE)
data$Timestamp <- dmy_hms(gsub(' GPS', '', data$Timestamp))
# Filter for relevant animal loggers
animal_loggers <- c(347, 350, 367, 370, 371, 383, 405, 411, 412, 416, 420, 422, 425, 2010, 2020, 2030, 2040, 2060, 2070, 2110, 2160)
filtered_data <- data %>%
filter(Receiver.ID %in% animal_loggers & Sender.ID %in% animal_loggers)
# Group data by 15-minute windows
grouped_data <- filtered_data %>%
mutate(Timestamp = floor_date(Timestamp, unit = "15 minutes")) %>%
group_by(Timestamp, Receiver.ID, Sender.ID) %>%
summarise(RSSI = mean(RSSI, na.rm = TRUE)) %>%
ungroup()
Creating RSSI Matrices
Next, we will create matrices that represent the RSSI values between different loggers over time.
Python Example
# Create matrices from the grouped data
def create_matrix(grouped_df):
unique_ids = sorted(set(grouped_df['Receiver ID']).union(set(grouped_df['Sender ID'])))
matrices = {}
for time, group in grouped_df.groupby(grouped_df['Timestamp'].dt.floor('15min')):
matrix = pd.DataFrame(index=unique_ids, columns=unique_ids, data=np.nan)
for _, row in group.iterrows():
matrix.at[int(row['Receiver ID']), int(row['Sender ID'])] = row['RSSI']
matrices[time] = matrix.replace(0, -np.inf)
return matrices
matrices = create_matrix(grouped_data)R Example
# Create matrices from the grouped data
create_matrix <- function(grouped_df) {
unique_ids <- sort(unique(c(grouped_df$Receiver.ID, grouped_df$Sender.ID)))
matrices <- list()
grouped_times <- split(grouped_df, grouped_df$Timestamp)
for (time in names(grouped_times)) {
group <- grouped_times[[time]]
matrix <- matrix(NA, nrow = length(unique_ids), ncol = length(unique_ids), dimnames = list(unique_ids, unique_ids))
for (row in 1:nrow(group)) {
matrix[as.character(group[row, 'Receiver.ID']), as.character(group[row, 'Sender.ID'])] <- group[row, 'RSSI']
}
matrices[[time]] <- matrix
}
return(matrices)
}
matrices <- create_matrix(grouped_data)Applying DBSCAN
We will apply DBSCAN to each matrix to identify clusters of animals based on their proximity.
Python Example
# Apply DBSCAN to each matrix and store clusters
def apply_dbscan(matrix):
coords = np.array([(i, j) for i in range(matrix.shape[0]) for j in range(matrix.shape[1]) if not np.isnan(matrix.iat[i, j])])
if len(coords) == 0:
return np.full(matrix.shape, -1) # Return a matrix full of -1 (indicating noise) if there are no coordinates
values = np.array([matrix.iat[i, j] for i, j in coords])
clustering = DBSCAN(eps=100, min_samples=2).fit(coords, sample_weight=values)
labels = np.full(matrix.shape, -1) # Default to noise (-1)
for (i, j), label in zip(coords, clustering.labels_):
labels[i, j] = label
return labels
# Apply DBSCAN to each matrix
clustered_matrices = {time: apply_dbscan(matrix) for time, matrix in matrices.items()}
R Example
library(dbscan)
# Apply DBSCAN to each matrix and store clusters
apply_dbscan <- function(matrix) {
coords <- which(!is.na(matrix), arr.ind = TRUE)
if (nrow(coords) == 0) {
return(matrix(-1, nrow(matrix), ncol(matrix))) # Return a matrix full of -1 (indicating noise) if there are no coordinates
}
values <- matrix[coords]
clustering <- dbscan(coords, eps = 100, minPts = 2, weights = values)
labels <- matrix(-1, nrow(matrix), ncol(matrix)) # Default to noise (-1)
labels[coords] <- clustering$cluster
return(labels)
}
# Apply DBSCAN to each matrix
clustered_matrices <- lapply(matrices, apply_dbscan)Visualizing Clusters
Finally, we will visualize the clusters using a heatmap and a slider to navigate through time.
Python Example
# Function to plot matrices with clusters using a slider
def plot_matrices_with_clusters(matrices):
times = list(matrices.keys())
initial_time = times[0]
unique_ids = matrices[initial_time].index
fig, ax = plt.subplots(figsize=(12, 8))
plt.subplots_adjust(bottom=0.25)
# Apply DBSCAN to the initial matrix
clusters = apply_dbscan(matrices[initial_time])
# Plot initial matrix with clusters
heatmap = sns.heatmap(matrices[initial_time], annot=clusters, fmt='', cmap='viridis', ax=ax)
cbar = heatmap.collections[0].colorbar
xticks = range(0, len(unique_ids), max(1, len(unique_ids) // 20))
yticks = range(0, len(unique_ids), max(1, len(unique_ids) // 20))
ax.set_xticks(xticks)
ax.set_xticklabels([unique_ids[i] for i in xticks], rotation=90)
ax.set_yticks(yticks)
ax.set_yticklabels([unique_ids[i] for i in yticks])
ax_time = plt.axes([0.2, 0.1, 0.65, 0.03], facecolor='lightgoldenrodyellow')
time_slider = Slider(ax_time, 'Time', 0, len(times)-1, valinit=0, valfmt='%0.0f')
def update(val):
time_idx = int(val)
current_time = times[time_idx]
# Apply DBSCAN to the current matrix
clusters = apply_dbscan(matrices[current_time])
# Update heatmap with new data and clusters
ax.clear()
sns.heatmap(matrices[current_time], annot=clusters, fmt='', cmap='viridis', ax=ax, cbar=False)
xticks = range(0, len(unique_ids), max(1, len(unique_ids) // 20))
yticks = range(0, len(unique_ids), max(1, len(unique_ids) // 20))
ax.set_xticks(xticks)
ax.set_xticklabels([unique_ids[i] for i in xticks], rotation=90)
ax.set_yticks(yticks)
ax.set_yticklabels([unique_ids[i] for i in yticks])
plt.draw()
time_slider.on_changed(update)
plt.show()
# Main function
def main():
# Plot matrices with clusters and slider
plot_matrices_with_clusters(matrices)
if __name__ == "__main__":
main()
R Example
library(ggplot2)
library(gganimate)
library(transformr)
# Function to create a heatmap plot
plot_heatmap <- function(matrix, title) {
melted <- as.data.frame(as.table(matrix))
names(melted) <- c("Receiver.ID", "Sender.ID", "RSSI")
ggplot(melted, aes(x = Receiver.ID, y = Sender.ID, fill = RSSI)) +
geom_tile() +
scale_fill_viridis_c() +
ggtitle(title) +
theme(axis.text.x = element_text(angle = 90, hjust = 1))
}
# Plot and animate the matrices over time
animate_matrices <- function(matrices) {
plots <- lapply(names(matrices), function(time) {
plot_heatmap(matrices[[time]], paste("Time:", time))
})
anim <- do.call(gganimate::transition_states, c(list(states = names(matrices), transition_length = 2, state_length = 1), plots))
anim_save("animal_behavior_clusters.gif", anim)
}
# Main function
main <- function() {
# Animate matrices with clusters
animate_matrices(matrices)
}
main()We can visualise data by using multidimensional scaling (MDS) to visualise the matrix of RSSI’s in between loggers in a 2D fashion. If we then run DBScan on the RSSI-data, and visualise the points alongside the MDS-visualization we see that the clustering makes sense and that DBScan can be used to identify clusters of animals.
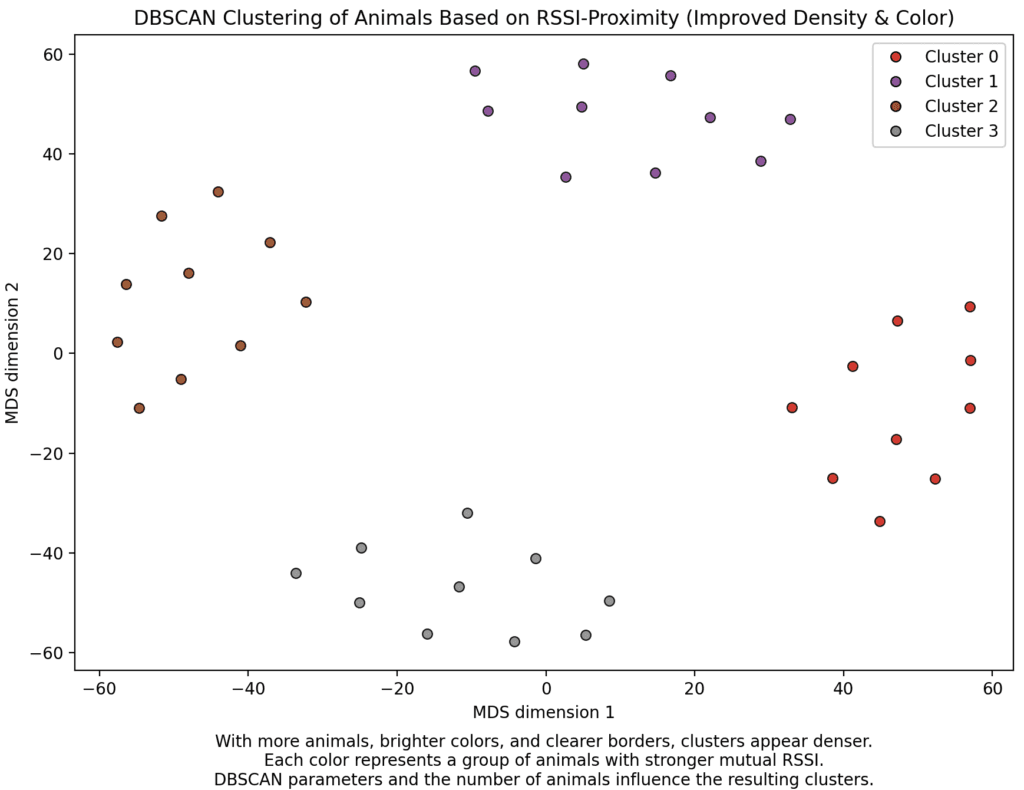
Conclusion
In this blog post, we demonstrated how to use DBSCAN to analyze animal behavioral data using proximity logger data. We walked through the data preparation, matrix creation, clustering with DBSCAN, and visualization of the results. By following these steps, you can gain valuable insights into animal interactions and behaviors over time. Both Python and R examples were provided to cater to different preferences and toolsets.